Came across this paper using complete mitochondrial genomes and all of the markers contained therein to assess Primate evolution.
A Mitogenomic Phylogeny of Living Primates
July 16, 2013
From the results and discussion:
We produced complete mt genome sequences from 32 primate individuals. From each individual, we obtained an average of 1508 tagged reads with an average length of 235 bp, yielding approximately 356 kb of sequence data corresponding to 21-fold coverage. All newly sequenced mt genomes had lengths typical for primates (16,280–16,936 bp;
Table S1), but the GC-content varied largely among taxa (37.78–46.32%,
Table S2,
Figure S1). All newly generated mt genomes consisted of 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes and the control region in the order typical for mammals. By combining the 32 newly generated data with 51 additional primate mt genomes, the dataset represents all 16 primate families, 57 of the 78 recognized genera and 78 of the 480 currently recognized species [
31].
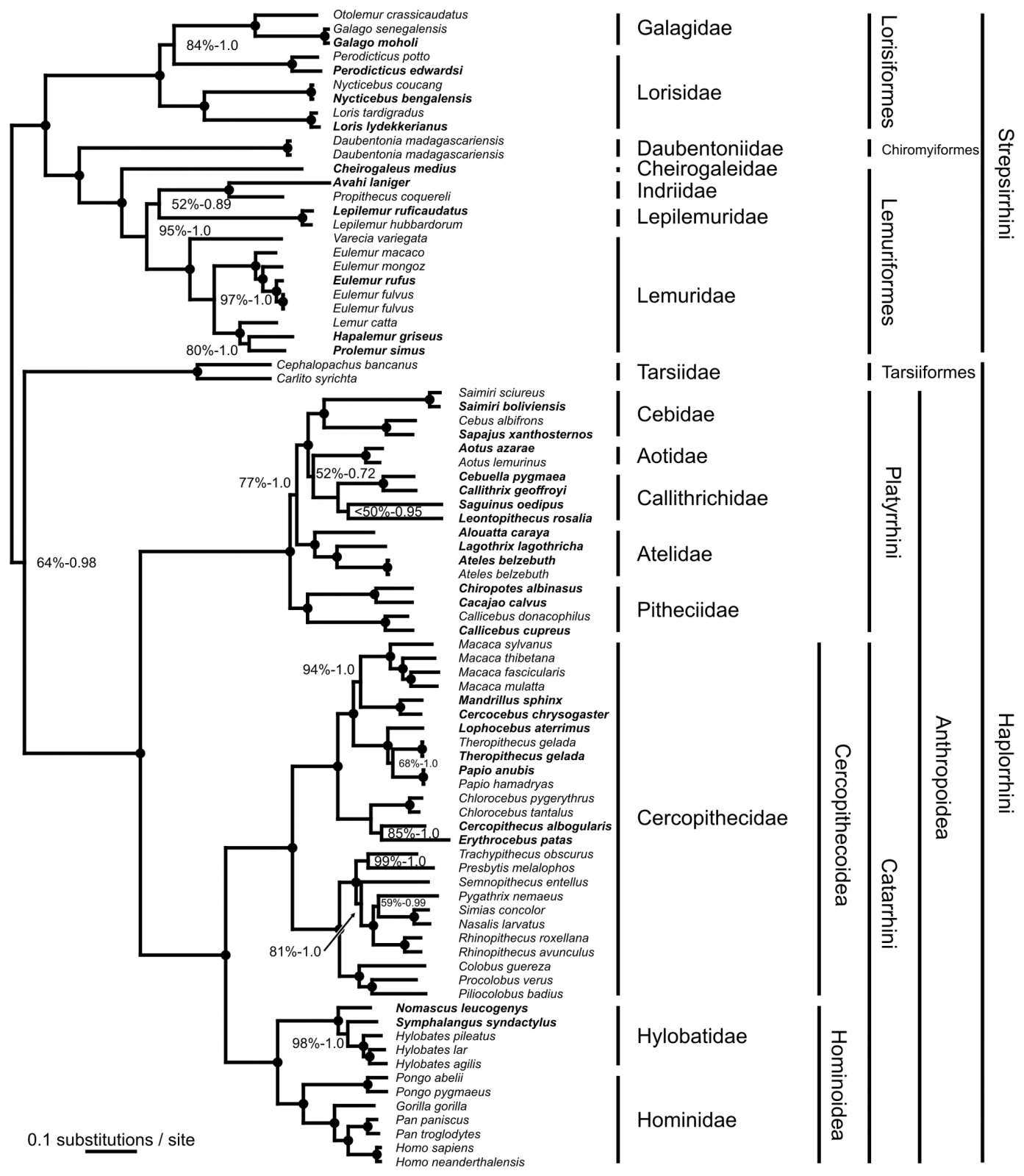
They used 81 complete mitochondrial genomes from primates representing all 16 families. The descriptions of the genomic content represent all of the markers that one could hope for. The use of these markers allow for the tracing of the ancestry of all of the primate taxa used, as shown in this phylogenetic tree, and such trees are produced as the output of a rigorous analysis - the same sort employed in the Canid paper.
Note that this includes humans, Neanderthals, etc.
This phylogenetic tree incorporates the tracing of mtDNA snps and other such markers. The shared ancestry of all Primates is thus proven.
The type of data used and the means of analysis employed have been BY YOU, so there is no denying the shared ancestry of human, chimps and other primates.
1 point.
1 source.
Explanation provided.